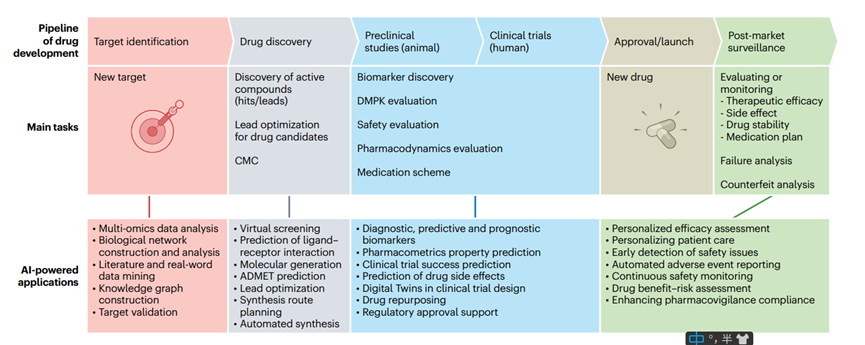
New Opportunities for PROTAC
Abstract
PROTAC (PROteolysis TArgeting Chimera, Protein Degradation Targeting Chimera) is a bifunctional molecule with a segment of ligand that binds the target protein and a segment of ligand that binds the E3 ubiquitin ligase at the other end, linked by a chain. In vivo, it can pull the target protein and E3 enzyme together, so that the target protein is tagged with ubiquitin and then degraded via the ubiquitin-proteasome pathway.
Mechanism of action
In vivo, PROTAC utilizes the ubiquitin-proteasome pathway to degrade target proteins as follows: the ligands at both ends of PROTAC recognize the E3 ubiquitin ligase and the target protein, forming a target protein-PROTAC-E3 ligase triplet complex that pulls in the target protein and the E3 ligase, which subsequently transfers ubiquitin to the target protein. The proteasome accepts the ubiquitin-tagged protein and cleaves it into a short peptide of 7-9 amino acids. After the target protein is degraded, the PROTAC molecule can be released to participate in the next protein degradation process, so this degradation has a catalytic effect, and a small dose of the drug can achieve efficient degradation.
Advantages of PROTAC
Unlike the mechanism of action of traditional small molecule inhibitors, PROTAC inhibits the degradation of target proteins by binding to the active site of the target protein, thus achieving the effect of disease treatment. Compared to it, PROTAC has many advantages.
Target site of action
Traditional small molecule inhibitors need to act on the active site of the target protein to inhibit its function, and this target protein accounts for less than 20% of disease-related proteins, while PROTAC technology only needs to pull in the target protein with E3 ubiquitin ligase to degrade the substrate protein, which can be applied to some transcription factors and other non-druggable targets.
Catalytic protein degradation function
Small molecule inhibitors act on target proteins to inhibit the activity and do not degrade them away; whereas PROTAC molecules can directly catalyze the degradation of target proteins.
Overcoming drug resistance
The action of small molecule inhibitors on target proteins may cause overexpression or mutation of target proteins, which may lead to drug resistance and make small molecule inhibitors lose their inhibitory function on target proteins; while PROTAC molecules degrade the target proteins through proteasome pathway, which can avoid drug resistance due to point mutation to a certain extent.
High selectivity
Nathanael S. Gray’s group had designed a small molecule inhibitor that can act on multiple proteins of the CDK family as the PROTAC molecule, which can selectively degrade CDK9 while not affecting other targets. This study illustrates that due to the synergistic interaction between the target protein and E3 ubiquitin ligase, the PROTAC molecule can provide selectivity on top of the small molecule inhibitor.
Low dosage and low toxicity
PROTAC molecules can directly catalyze the degradation of target proteins, and after the target proteins are degraded, they can be released to participate in the next protein degradation process, achieving significant and long-lasting effects at low doses.
ARV-825
ARV-825 is a specific-derived bifunctional protein hydrolysis-targeted chimera (PROTAC) that recruits BRD4 to the E3 ubiquitin ligase cereblon. ARV-825 actively recruits BRD4 into brain cells, leading to rapid and efficient degradation of the former via the proteasome. Given that the Kds of the BRD4 and cerebrospinal binding fractions in ARV-825 are 28-90 nM and ~3 μM, respectively, this suggests that ARV-825 acts in a subchemotropic manner to mediate BRD4 degradation. Compared to BRD4 inhibitors, ARV-825 treatment resulted in prolonged BRD4 downregulation and downstream signaling inhibition.
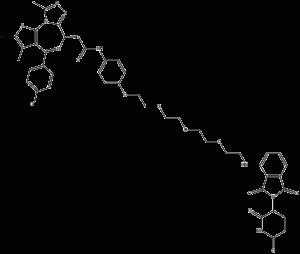
ARV-825 structural formula
MZ-1
MZ-1 is a bromodomain-containingprotein4 (BRD4) PROTAC degrader.MZ-1 binds to the Brd structural domains Brd2BD1, BChemicalbookrd2BD2, Brd3BD1, Brd3BD2, Brd4BD1 and Brd4BD2, corresponding to the Kd values of 62nM, 60nM, 21nM, 13nM, 39nM and 15nM, respectively.
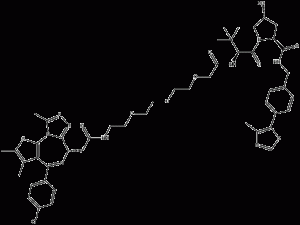
MZ-1 structural formula
dBET6
dBET6 is a BET bromodomainsPROTAC degradation agent with good cellular permeability and an IC50 value of 14 nM for binding to BRD4. dBET6 is effective in most cancer cell lines. dBET6 has a significant degradation effect in cells at concentrations below the molar level. dBET6 also induces c-MYC downregulation and apoptosis. In addition, dBET6 was well tolerated. In a diffuse T-ALL mouse model, dBET6 significantly reduced leukemic load.
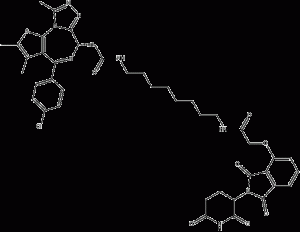
dBET6 structural formula
ACBI1
ACBI1 is a potent, PROTAC technology-based degrader of the BAFATPase subunits SMARCA2 and SMARCA4, and a degrader of the PBAF member PBRM1, with DC50 values of 6nM, 11nM, and 32nM for SMARCA2, SMARCA4 and PBRM1, respectively.ACBI1 consists of a bromodomain ligand, linker, and E3 ubiquitin ligase VHL, which induces antiproliferative effects and apoptosis.
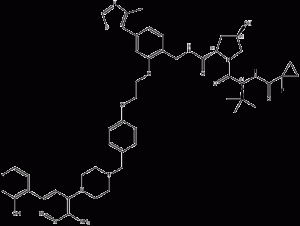
ACBI1 structural formula
ARV-771
ARV-771 is a potent pan-(bromodomainandextra-terminal) BET degrader, a novel BET-PROTAC (proteolysis-targetingchimChemicalbookera), for BRD2(1), BRD2(2), BRD3(1), BRD3(2), BRD4(1) and BRD4(2) with Kd values of 34nM, 4.7nM, 8.3nM, 7.6nM, 9.6nM and 7.6nM, respectively.
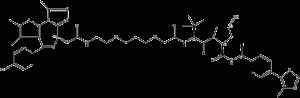
ARV-771 structural formula
dBET1
dBET1 is a CRBN-based BET degrader with high selectivity for BRD4 with an IC50 of 20 nM. Among the 7429 Chemicalbook detected proteins, only the expression of tumor proteins MYC, PIM1 BRD2, BRD3, and BRD4 were significantly decreased by dBET1 treatment.
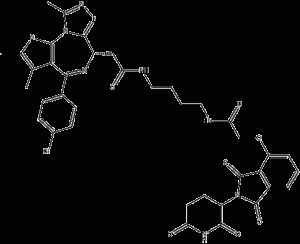
dBET1 structural formula
MD-224
MD-224 is a highly efficient small molecule (MDM2) degradation agent based on protein hydrolysis localization chimeras (PROTAC).MD-224 consists of Cereblon and MDM2 ligands.MD-224 induced rapid degradation of MDM2 (<1 nM) in human leukemia cells and inhibited RS4;11 cell growth with an IC50 value of 1.5 nM. 224 has the potential to be a new class of anticancer agents.
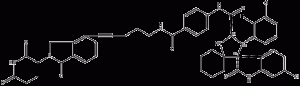
MD-224 structural formula
SD-36
SD-36 is a PROTAC degradation agent of STAT3 protein. It efficiently induces STAT3 protein degradation in vitro and in vivo and exhibits a higher selectivity than other STAT members. Induction of STAT3 degradation leads to a strong inhibition of its transcriptional network Chemicalbook in leukemia and lymphoma cells.SD-36 inhibits acute myeloid leukemia and mesenchymal large-cell growth of subpopulations of lymphoma cell lines. It achieved complete and durable tumor regression in multiple xenograft mouse models with well-tolerated dosing regimens.
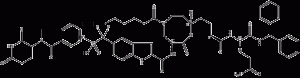
SD-36 structural formula
BSJ-03-123
BSJ-03-123 is a PROTAC linked by Cereblon and CDK ligand, an effective, novel CDK6-selective small molecule degradation agent.
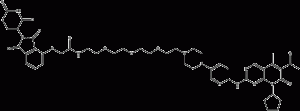
BSJ-03-123 structural formula
GMB-475
GMB-475 is a PROTAC-based Bcr-Abl1 tyrosine kinase degrader that overcomes Bcr-Abl1-dependent drug resistance. GMB-475 targets the Bcr-Abl1 protein and recruits the E3 Von Hippel Lindau (VHL) ligase. Leads to ubiquitination and subsequent degradation of oncogenic fusion proteins.
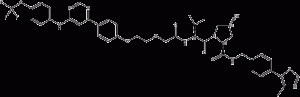
GMB-475 structural formula
β-NF-JQ1
β-NF-JQ1 is a PROTAC that recruits the Aryl Hydrocarbon Receptor E3 ligase to target proteins. β-NF-JQ1 uses β-NF as an AhR ligand to target bromodomain (BRD)-containing proteins, induces AhR and BRD protein interactions, and shows potent anticancer activity associated with protein knockdown activity. activity.
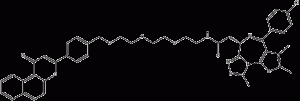
β-NF-JQ1 structural formula
TL13-12
TL13-12 is a potent ALK-PROTAC degradant that inhibits ALK activity with an IC50 value of 0.69 nM and promotes the degradation of other kinases, including Aurora A, FER, PTK2 and RPS6KA1 with IC50 values of 13.5 nM, 5.74 nM, 18.4 nM, and 65 nM, respectively.TL13-12 is composed of TAE684 (HY-10192) and Cereblon ligand Pomalidomide (HY-10984).

TL13-12 structural formula
TL13-112
TL13-112 is a potent and selective ALK-PROTAC degradation agent that inhibits ALK activity with an IC50 value of 0.14 nM and promotes degradation of other kinases including Aurora A, FER, PTK2, and RPS6KA1 with IC50 values of 8550 nM, 42.4 nM, 25.4 nM, and 677 nM, respectively.TL13 -112 is bound by Ceritinib (HY-15656) and Cereblon ligand Pomalidomide (HY-10984).
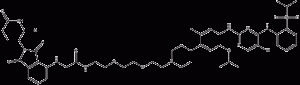
TL13-112 structural formula
Summary of Prospects
More than 80% of proteins in human cells lack enzymatic activity or drug-forming sites on their surfaces, making most small-molecule drugs or monoclonal antibodies ineffective, while PROTAC molecules can capture and degrade them for clearance. PROTACs can be studied not only for classical drug targets (estrogen receptors, protein kinases, etc.) but also for transcription factors, protein backbones, etc. In the future, it holds promise to play a role in the treatment of multiple diseases, especially in the direction of cancer.
The release of global PROTAC drug-human trial data is a milestone event in translating PROTAC technology into new treatment modalities to help patients. As more and more PROTAC molecules enter the clinical or preclinical study phase, it is believed that the technology will develop rapidly in the coming years.
Reference:
- Cruz Walma DA, Chen Z, Bullock AN, Yamada KM. Ubiquitin ligases: guardians of mammalian development. Nat Rev Mol Cell Biol. 2022 May;23(5):350-367. doi: 10.1038/s41580-021-00448-5. Epub 2022 Jan 25. PMID: 35079164.
- Cao C, He M, Wang L, He Y, Rao Y. Chemistries of bifunctional PROTAC degraders. Chem Soc Rev. 2022 Aug 15;51(16):7066-7114. doi: 10.1039/d2cs00220e. PMID: 35916511.
- Wang C, Zheng C, Wang H, Zhang L, Liu Z, Xu P. The state of the art of PROTAC technologies for drug discovery. Eur J Med Chem. 2022 May 5;235:114290. doi: 10.1016/j.ejmech.2022.114290. Epub 2022 Mar 15. PMID: 35307618.
- Qi SM, Dong J, Xu ZY, Cheng XD, Zhang WD, Qin JJ. PROTAC: An Effective Targeted Protein Degradation Strategy for Cancer Therapy. Front Pharmacol. 2021 May 7;12:692574. doi: 10.3389/fphar.2021.692574. PMID: 34025443; PMCID: PMC8138175.
- Lee J, Lee Y, Jung YM, Park JH, Yoo HS, Park J. Discovery of E3 Ligase Ligands for Target Protein Degradation. Molecules. 2022 Oct 2;27(19):6515. doi: 10.3390/molecules27196515. PMID: 36235052; PMCID: PMC9573645.
- Duan W, Yu M, Chen J. BRD4: New hope in the battle against glioblastoma. Pharmacol Res. 2023 May;191:106767. doi: 10.1016/j.phrs.2023.106767. Epub 2023 Apr 13. PMID: 37061146.